Biomedical knowledge graphs
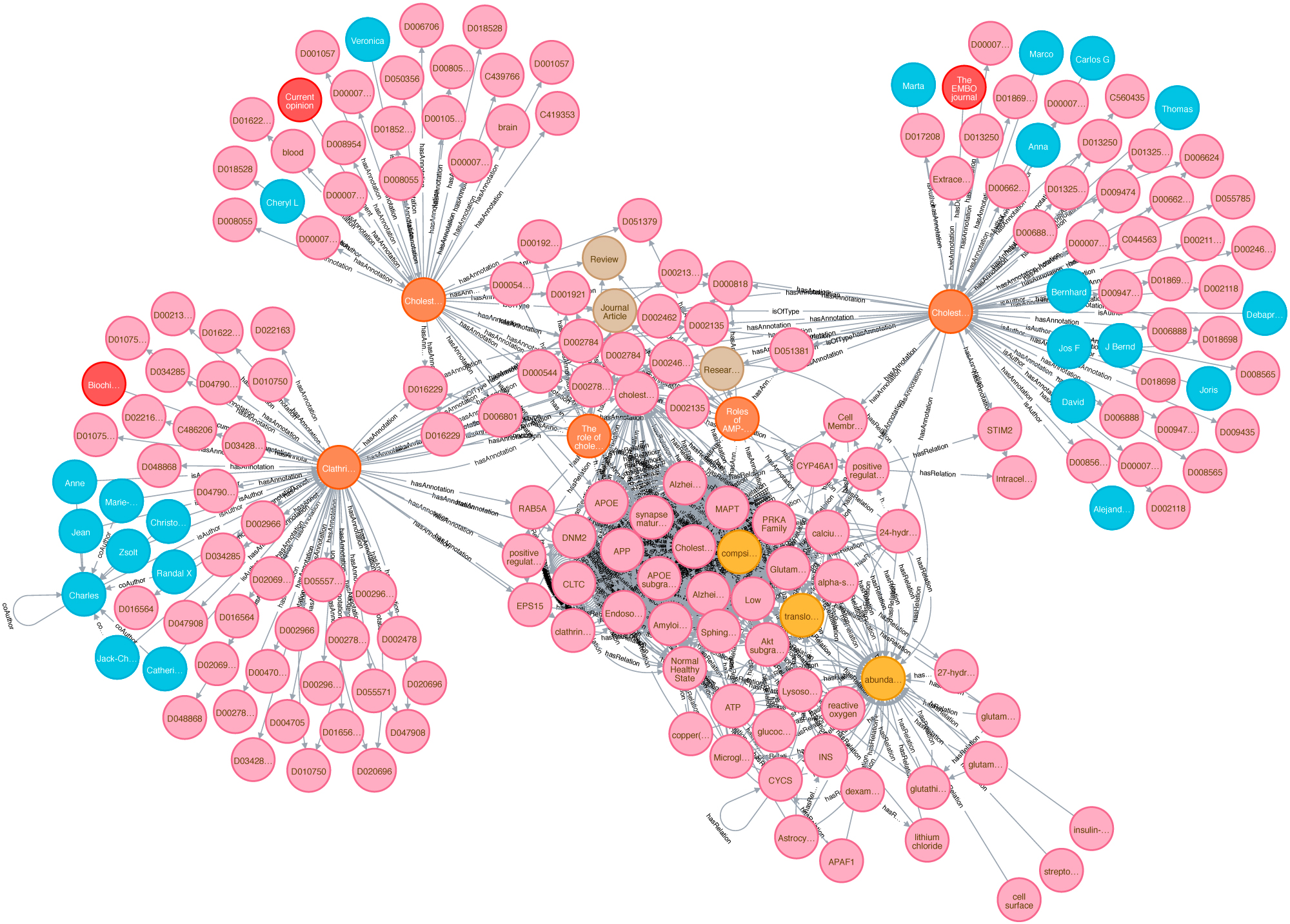
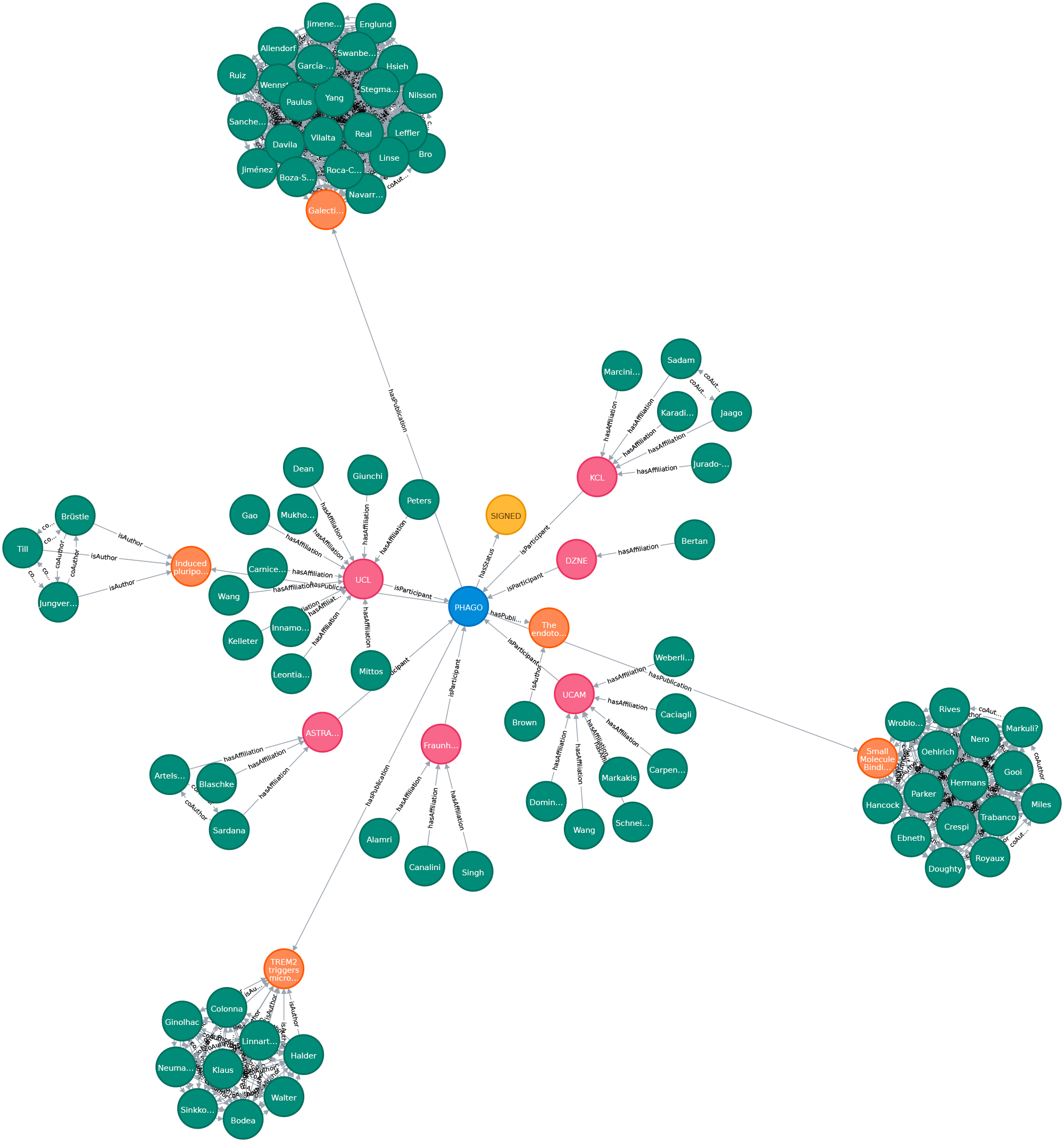
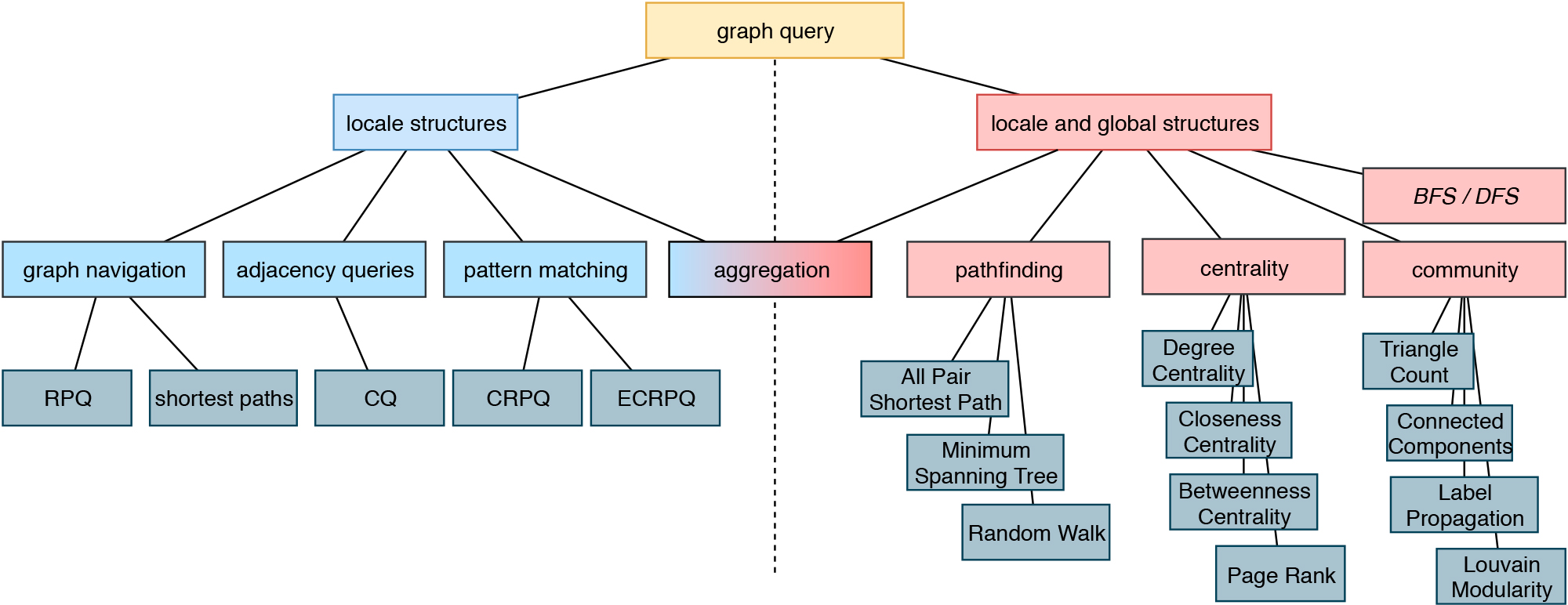
Biomedical knowledge graphs play a central role in big data integration. Bringing unstructured text into a structured, comparable format is one of the key assets. As cause and effect models, knowledge graphs can potentially facilitate clinical decision making or help to drive research towards precision medicine. Data and Knowledge Management, sometimes also called Information Management, is a core topic of Data Science. It is also a interdisciplinary field touching economics (how efficient and expensive is the solution?), psychology (do people use this solution in a way that was intended?) and of course computer science. Our aim is to build sustainable data infrastructure for biomedical data, personalized medicine, drug repurposing, reproducible AI and knowledge discovery.
A »knowledge graph« (sometimes also called a semantic network) is a systematic way to connect information and data points to knowledge. It is thus a crucial concept on the way to generate knowledge and wisdom, to search within data, information and knowledge. However, the power of knowledge graphs critically depends on context information and data integration. Here we provide a novel semantic approach towards a context enriched biomedical knowledge graph utilizing data integration with linked data. This graph concept can be used for graph embedding applied in different approaches, e.g with focus on topic detection and knowledge discovery. Thus, connecting knowledge graphs with context is a key feature. In this project we want to establish a novel systematic approach to knowledge discovery using contexts in knowledge graphs. For this, we enrich the existing graph structures and build a context hypergraph.
We create a proof-of-concept giant knowledge graph using labeled property graphs to test graph algorithms and provide a feasible environment to apply semantic graph embeddings. It is a highly scalable cloud-based service environment. This dense large scale labeled property graph testing system currently holds more then 75M nodes and 960M edges. The basis for generating our large-scale Knowledge Graph representation is biomedical literature (e.g.from PubMed and PMC). We also integrated bibliographic data and metadata from DBLP, monthly snapshot release of December 2019, see here. Since the basic data coming from SCAIView is already annotated with different biomedical ontologies, we decided to annotate CSO to DBLP data. We enriched our graph with data from the EU Open Data Portal (CORDIS - EU research projects under Horizon 2020). This data set is free to reuse for both commercial or non-commercial purpose. Here, we integrated projects, their status, affiliations, persons and authors of publications mentioned in their data set.
Download subset of PHAGO Graph
This is the basis for answering semantic questions, graph queries and extensions based on NLP, Text Mining, FAIR Data and a step towards reproducible AI. This graph allows to compare research data records from different sources as well as the selection of relevant data sets using graph-theoretical algorithms.
Medical Knowledge Space
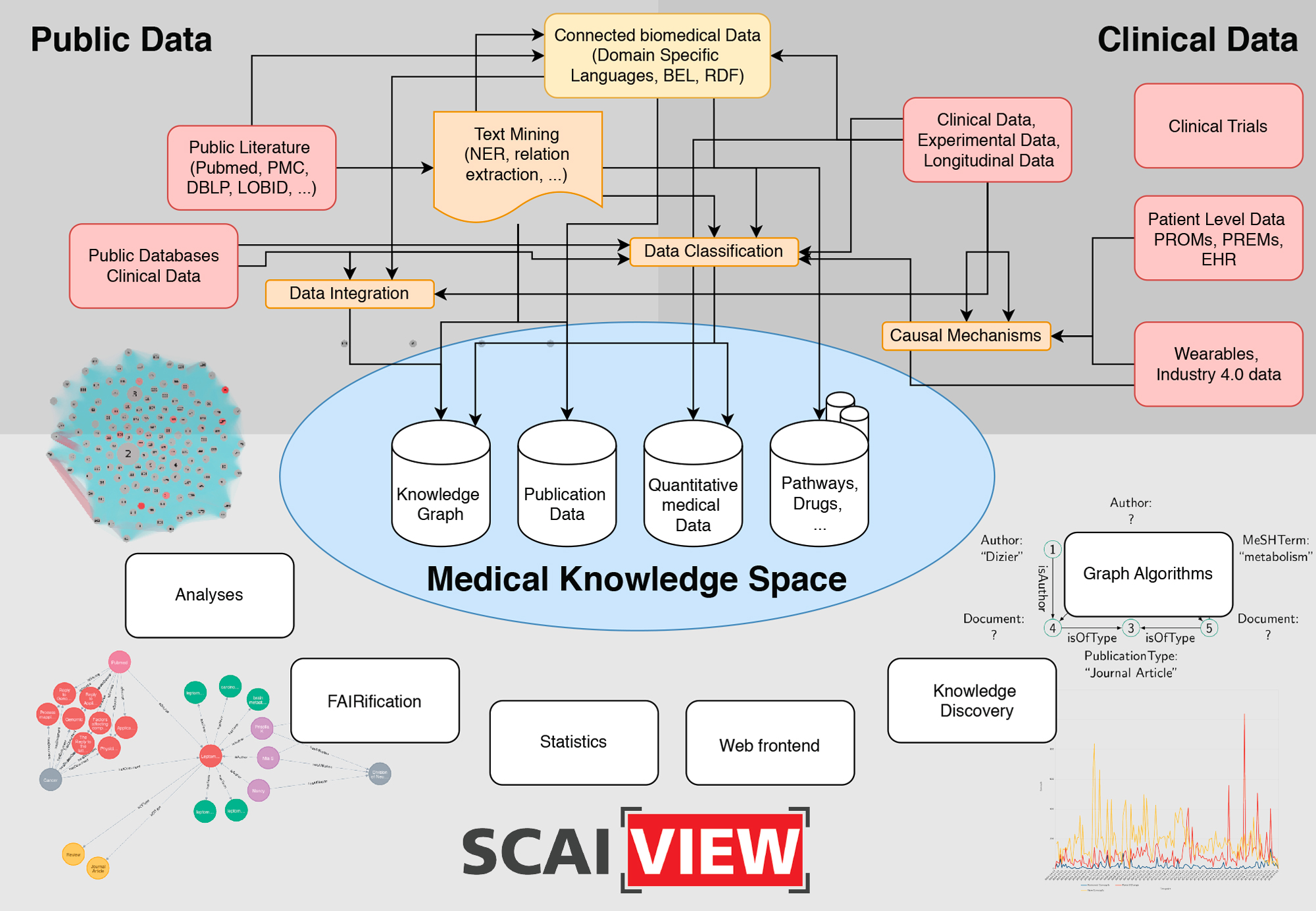
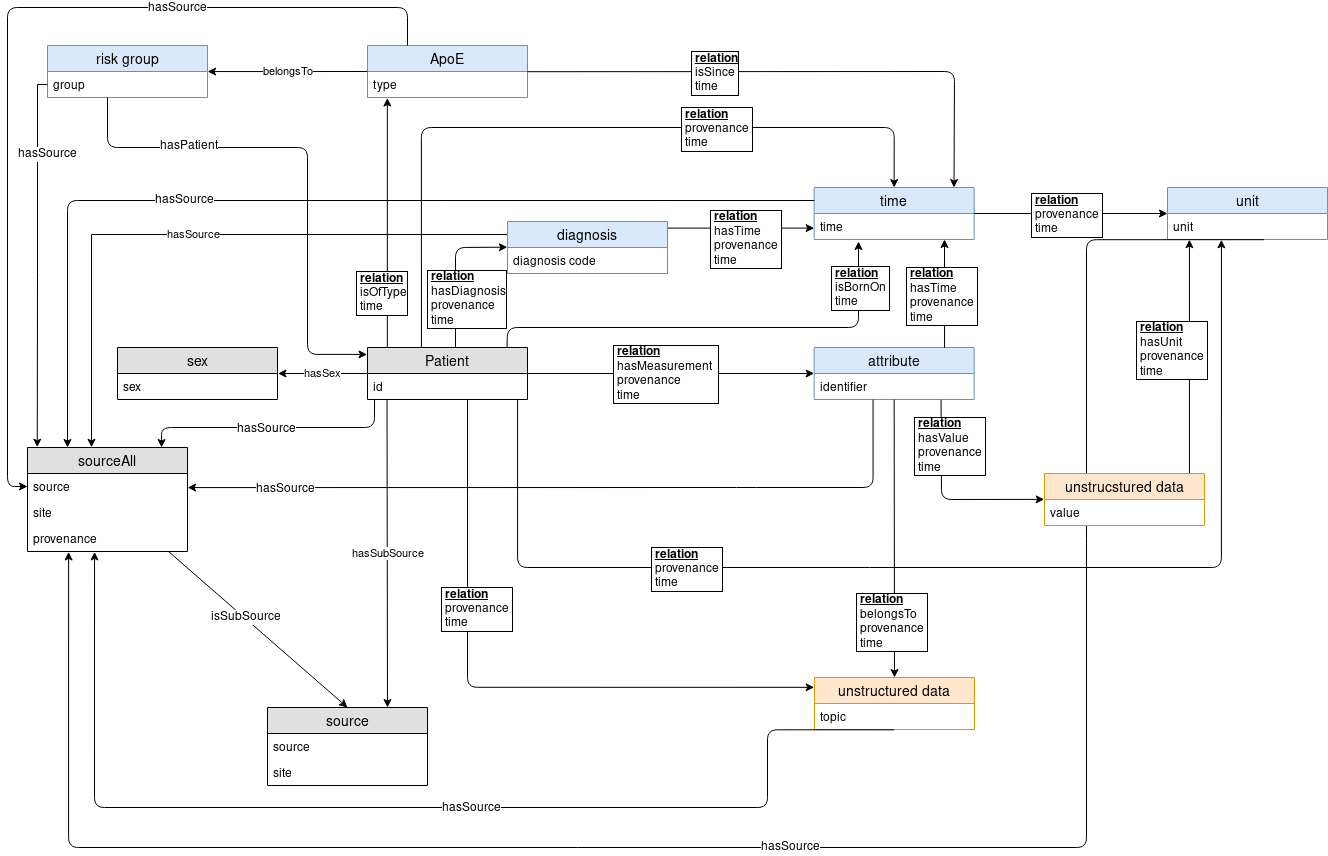
The Medical Knowledge Space is an appropriate place for clinical and medical data, both from public sources as well as clinical and patient-level data. It provides a systematic literature search and a specific analysis of available clinical data. This helps to identify the most relevant functional domains, which will be developed into big data approaches. The detailed analysis of the user needs as well as of the data involved can be carried out through an in-depth consulting and mutual learning progress combining all available data sources in an interoperability layer.
The overall concept underlying the SCAI.BIO Medical Knowledge Space is to bring analysis of medical knowledge directly to the data or patients in a patient-centric manner. We will do this by embracing the concept of scalability, where we develop tools and technologies that can reach patients at any geographic location. The huge advances in the development of health-care devices, including electronic health records, portal technologies, and wireless communications, will have a key role in future care. Of particular interest is for example the fact, that mobile devices may be used for realtime monitoring, including a variety of sensors to detect changes in the health status of the person.
The MKS provides methods for harmonization and interoperability between the services, the facilitation of their easy discovery, the sharing and usage of data sources as well as of the processing, analysis and modelling tools and finally a better documentation and reproducibility. By developing and subsequently providing the environment to move from raw data acquired in large samples, be it diagnosis data, molecular data or clinical information into the format required for advanced data analysis and modelling, the interoperability layer is a critical link between the clinical and modelling based projects in the current proposal. We provide several analyses tools and interfaces to apply graph queries and knowledge discovery. Whenever possible, we will provide FAIR data.
[1] Dörpinghaus, J. and A. Stefan. "Optimization of Retrieval Algorithms on Large Scale Knowledge Graphs." arXiv preprint arXiv:2002.03686 (2020).
[2] Dörpinghaus, Jens, et al. "Towards context in large scale biomedical knowledge graphs." arXiv preprint arXiv:2001.08392 (2020).
[3] Dörpinghaus, Jens, Carsten Düing, and Vera Weil. "A Minimum Set-Cover Problem with several constraints." 2019 Federated Conference on Computer Science and Information Systems (FedCSIS). IEEE, 2019.
[4] Dörpinghaus, Jens, and Andreas Stefan. "Knowledge Extraction and Applications utilizing Context Data in Knowledge Graphs." 2019 Federated Conference on Computer Science and Information Systems (FedCSIS). IEEE, 2019.
[5] Dörpinghaus, Jens, and Marc Jacobs. "Semantic Knowledge Graph Embeddings for biomedical Research: Data Integration using Linked Open Data." Proceedings of the Posters and Demos Track of the 15th International Conference on Semantic Systems – SEMANTiCS2019, 2019.
[6] Dörpinghaus, J. et al. “Context graph for biomedical research data: A FAIR and open approach towards reproducible research in Medicine”, 3rd Annual MAQC Society Conference 2019.
[7] Dörpinghaus, J. et al. "SCAIView–A Semantic Search Engine for Biomedical Research Utilizing a Microservice Architecture." Proceedings of the Posters and Demos Track of the 14th International Conference on Semantic Systems – SEMANTiCS2018, 2018