Johanna Driever
Transformer-based prediction of Parkinson’s disease using electronic health records
Master student Johanna Driever explains how she used a natural language processing model in combination with electronic health records to predict Parkinson’s disease.
Motivation
Disease prediction is an application of machine learning that relies on analyzing patterns from large data sets, such as electronic health records (EHRs), to identify diseases. Despite the promising potential of EHRs as an input data source for machine learning models, the irregularity of the data can present challenges for training accurate models. EHR data is an irregular discrete time series, making it challenging to use as input data for machine learning models. Doctor visits may not follow a predictable schedule, and there can be long gaps between observations. As a result, it can be difficult to identify data patterns relevant to disease prediction.
Implementing a transformer-based approach for the prediction of Parkinson's disease
Our study aims to build an ML predictor for Parkinson’s disease (PD), a neurodegenerative disease affecting the motor system. For our approach to predict PD, we used EHR sourced from UK Biobank to include the patient’s medical history and environmental factors as well as different Polygenic Risk Scores (PRS) for PD to include genetic information.
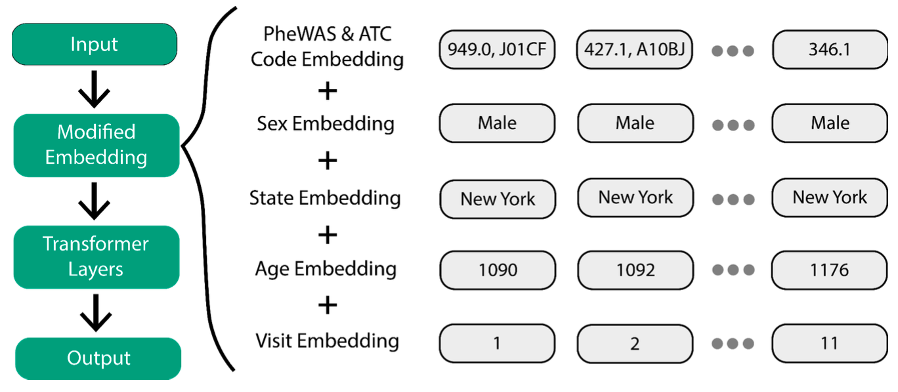
To overcome the challenge of using EHR data for disease prediction, we implemented a Transformer-based approach using SCAI’s previously published Ex-MED-BERT[1] model and two tree-based models, namely XGBoost and Random Forest. Ex-MED-BERT is an adaptation of BERT[2], a natural language processing model developed by Google. Before fine-tuning for a particular task, BERT first learns a representation of the input data on a large, unlabeled dataset. This pre-training step improves the performance of BERT on fine-tuning datasets, which is particularly important in medical applications. The EHR data is embedded with a modified embedding layer (Figure 1) that includes not only medical information but also chronological information, such as the patient's age at the time of the visit. After training the classifiers, explainable AI methods were employed to interpret the behavior of the ML models. First, a feature importance analysis was conducted, followed by the construction of a Bayesian Network (BN) on the 100 most important features to understand feature dependencies better.
The performances of the different approaches were similar, and all of them yielded acceptable results in terms of the model’s capabilities to predict PD. To improve the Ex-MED-BERT model further, we plan to pre-train it with the complete UK Biobank dataset so that it can learn a better representation of British data. Additionally, we aim to include more genetic information and lifestyle information, such as smoking, alcohol and caffeine consumption, and other environmental factors linked to PD. We hope that by further improving our model, we can better understand early PD prediction and contribute to a machine-learning-supported diagnostic procedure for PD.
References:
- Manuel Lentzen et al. “A Transformer-Based Model Trained on Large Scale Claims Data for Prediction of Severe COVID-19 Disease Progression”. In: medRxiv (2022). doi: 10.1101/2022.11.29.22282632.
- Jacob Devlin et al. “BERT: Pre-training of Deep Bidirectional Transformers for Language Understanding.” In: CoRR abs/1810.04805 (2018). arXiv: 1810.04805.