Yihao Wang
Multi-ontology embeddings approach on human-aligned multi-ontologies representation for gene-disease associations prediction
Master student Yihao Wang explains how he improved Natural Language Processing (NLP) models by infusing prior knowledge from ontologies.
Motivation
Knowledge graphs and ontologies are structured data that can capture relations between entities. They provide rich contextual knowledge in the biomedical domain and can be further employed for a variety of Natural Language Processing (NLP) downstream tasks, such as gene-disease association prediction. Instead of only using language models (e.g. BERT), we investigated whether knowledge enrichment with ontologies is a promising way to increase the predictive power of the language model.
Human-aligned multiple ontologies representation for gene-disease associations prediction
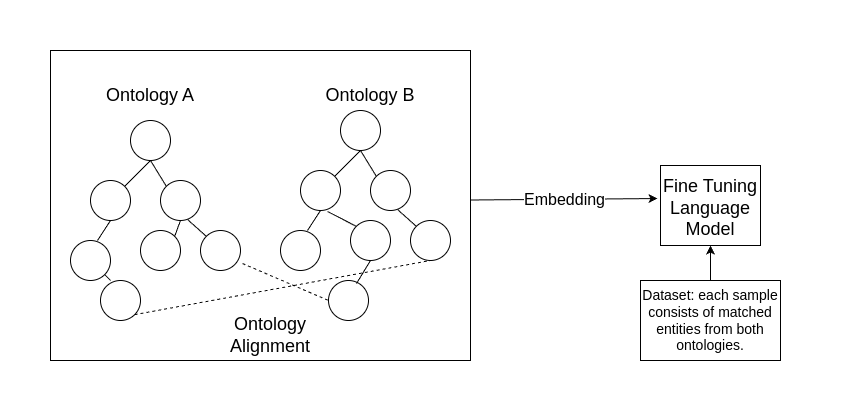
We used TBGA [1], a large-scale dataset of gene-disease associations, to benchmark and demonstrate our approach. We chose Human Disease Ontology (DOID) and Ontology of Genes and Genomes (OGG) as the knowledge enrichment, which covers all the genes and diseases appeared in the TBGA dataset. We combined two ontologies into one by manually adding "bridges". Bridges are gene-disease pairs with a high frequency of occurrence that we manually selected based on the dataset. The intention behind this pre-processing stage is that multiple ontologies from different domains require a reasonable spatial alignment for further embedding. The number of bridges is restricted since we want to keep the manual annotation as simple as possible and explore the generality of our approach for further researchers using different ontologies.
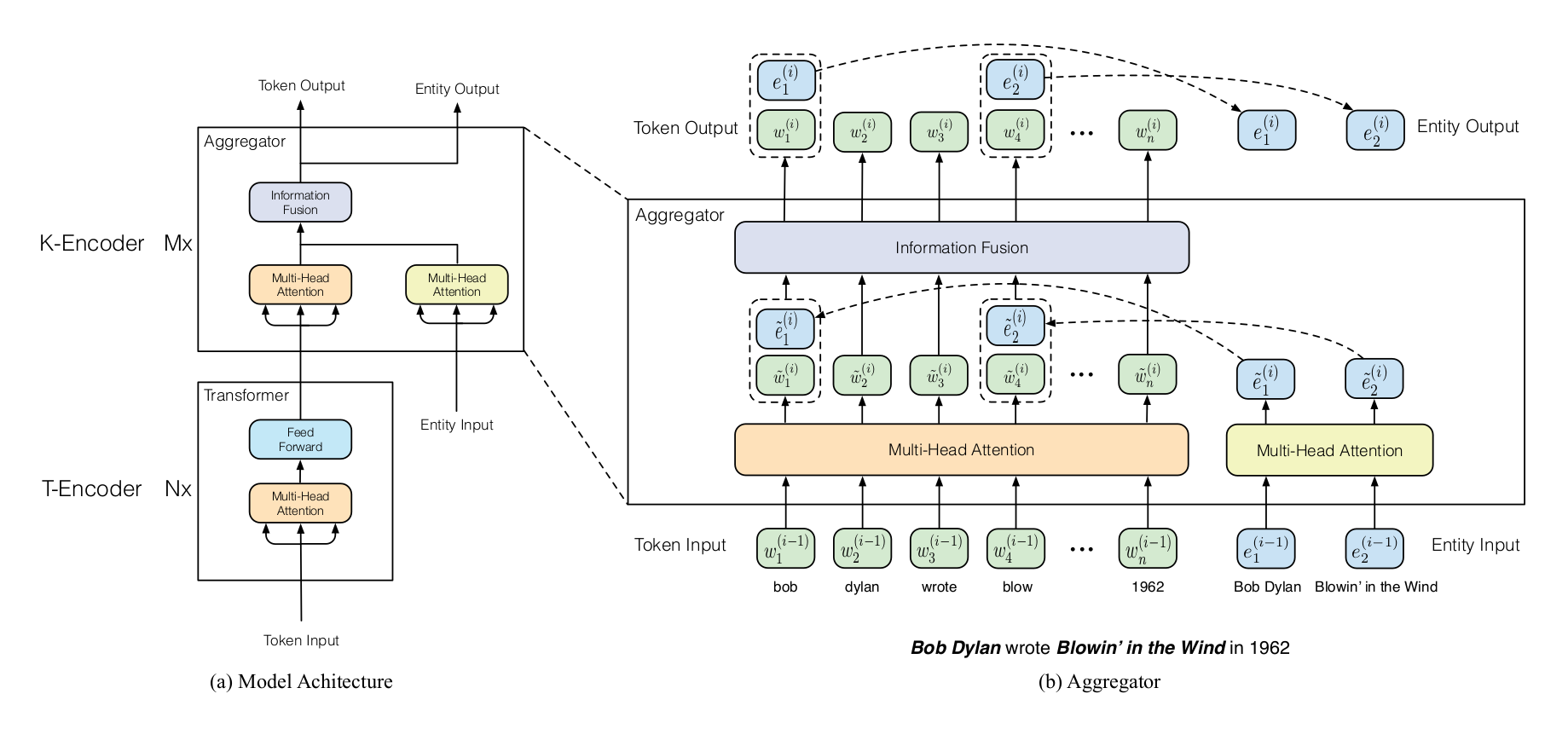
Furthermore, we used ERNIE [2], an enhanced language representation model, which is capable of infusing knowledge graphs as additional knowledge. We fine-tuned the ERNIE model by infusing the representations of multiple aligned ontologies. The experiments showed that the knowledge-enriched model outperforms BERT. Thus, this indicated that incorporating a small number of cross-references (bridges) across ontologies can enhance the performance of base models without the need for more complex and costly training. Furthermore, we expected that adding more bridges would bring further improvement based on the trend we observed in the experiments.
In conclusion, our work demonstrated the benefits of utilizing structured knowledge, i.e., embedding multiple aligned ontologies as prior knowledge in NLP tasks. Besides, the manual alignment of ontologies brought out in this work is constrained in reasonable complexity. The idea of our work to improve the language model's performance can be further adapted to other different tasks, e.g., multi-drug interaction, association of different psychiatric disorders, etc.
References:
- Marchesin, S., and Silvello, G. (2022). TBGA: a large-scale gene-disease association dataset for biomedical relation extraction. BMC bioinformatics, 23(1), 1-16.
- Zhang, Z., Han, X., Liu, Z., Jiang, X., Sun, M., & Liu, Q. (2019). ERNIE: Enhanced language representation with informative entities. arXiv preprint arXiv:1905.07129.