Diego Valderrama
Scientific machine learning for pharmacokinetics modeling
PhD student Diego Valderrama explains how hybrid models can be used as an interpretable alternative to black-box machine learning approaches for pharmacokinetics modeling.
Motivation
Prediction of pharmacokinetic (PK) effects is crucial in the drug discovery process. Since they determine the concentration in the body, the dosage to be used, and the anticipated rate of a drug's elimination in the plasma (also known as clearance) are crucial parameters during clinical studies. As a result, the main objective of PK prediction is to find clinical candidates who offer a concentration-time profile (PK profile) suitable for the effectiveness and safety of a particular medication.
The most used approach for PK prediction is implementing complex ordinary differential equations (ODE) systems, whose equations can sometimes be defined trivially and require a high degree of expertise. Additionally, their parameters and variability are highly dependent on specific covariates and are being learned via probabilistic methods, increasing the complexity of such approaches.
Black-box machine learning approaches – a half-baked solution
Although high-accuracy machine learning methods have been proposed for PK prediction, these approaches need to incorporate the natural physiological structure of the problem to be solved. Moreover, since most of these models are black-box approaches, they need to be interpretable, thus providing a half-baked solution. This is because although the PK profile is the ultimate expected output, it does not allow for the inference of essential factors like clearance.
Hybrid models – a promising idea for finding the perfect balance
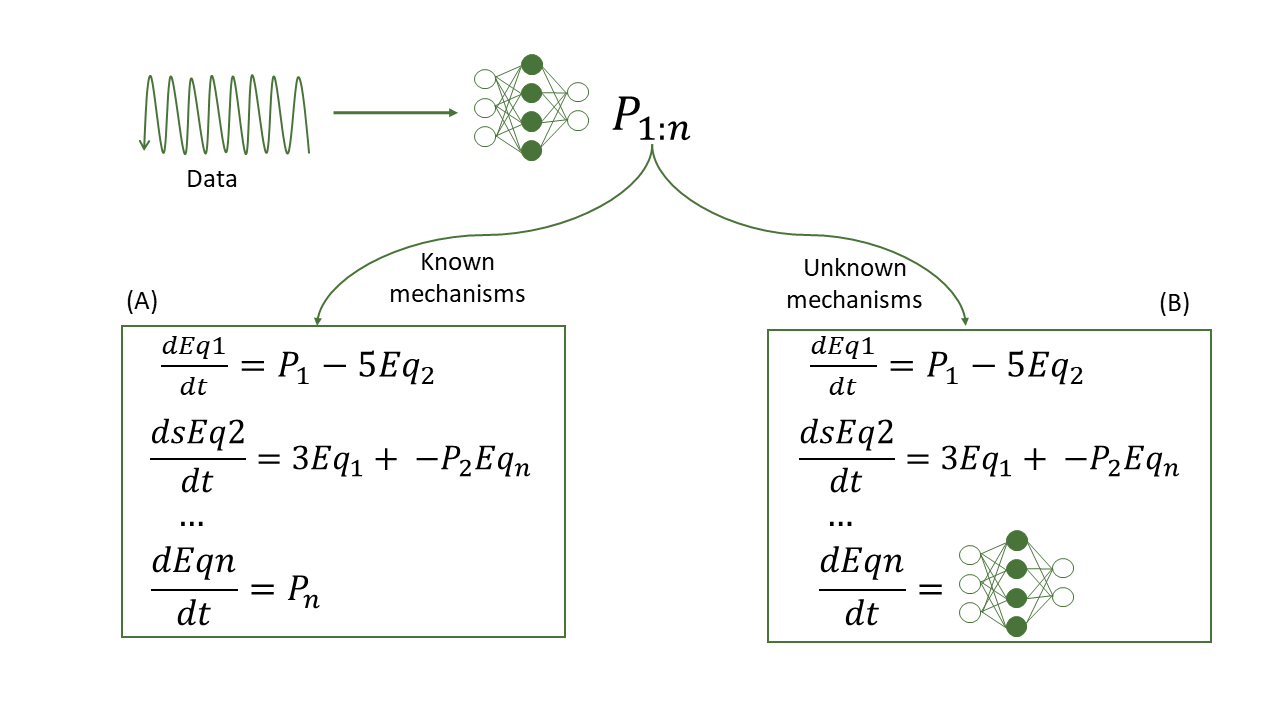
In this project, we are developing hybrid models following the scientific machine learning methodology. In essence, we combine mechanistic frameworks, specifically differential equations and machine learning, which lets us take advantage of both methodologies. The mechanical framework allows for including physiological constraints and domain expertise in equations. At the same time, a machine learning architecture helps manage multimodal (and potentially high dimensional) data, unknown mechanisms, and missing values. Additionally, the model gains interpretability thanks to an ODE system, which significantly benefits black box models.
Our research is currently concentrated on learning parameters (and their variability) that define an ODE system for particular medications, as well as models that involve some pure machine learning compartments, which are used when it is not straightforward to define the behavior in a compartment by a differential equation. We use the inverse problem approach to identify the optimal set of parameters without defining precise relationships between the parameters and the input data or using patient covariates as input.
References:
- Valderrama, D., Ponce‐Bobadilla, A. V., Mensing, S., Fröhlich, H., & Stodtmann, S. (2023). Integrating machine learning with pharmacokinetic models: Benefits of scientific machine learning in adding neural network components to existing PK models. CPT: Pharmacometrics & Systems Pharmacology.
- Reichel, A., & Lienau, P. (2016). Pharmacokinetics in drug discovery: an exposure-centred approach to optimising and predicting drug efficacy and safety. New approaches to drug discovery, 235-260
- Zou, P., Yu, Y., Zheng, N., Yang, Y., Paholak, H. J., Yu, L. X., & Sun, D. (2012). Applications of human pharmacokinetic prediction in first-in-human dose estimation. The AAPS journal, 14, 262-281.
- Negus, S. S., & Banks, M. L. (2018). Pharmacokinetic–pharmacodynamic (PKPD) analysis with drug discrimination. The Behavioral Neuroscience of Drug Discrimination, 245-259.