Zexin Li
Modeling of disease progression in Huntington’s disease
Zexin Li presents her work on data-driven disease subtyping, which is a promising approach to overcome these challenges in a mechanism-agnostic manner.
Estimating disease progression from an early stage is one of the challenging tasks in precision medicine, particularly for diseases with a high degree of variation across patients. Here, neurodegenerative diseases are a case in point since they are characterized by heterogeneous progression and multifaceted symptoms and have complex pathogenic mechanisms. In this post we take a look into data-driven disease subtyping, which is a promising approach to overcome these challenges in a mechanism-agnostic manner.
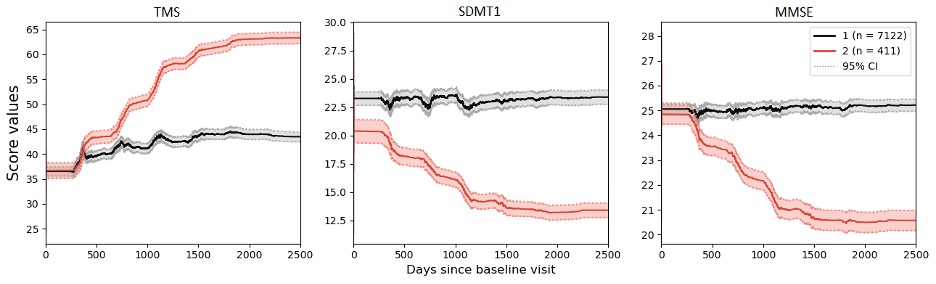
In our study, we leveraged a hybrid modeling approach to analyze a Huntington’s disease dataset (https://enroll-hd.org/) collected from worldwide observational research. To disentangle disease heterogeneity, we explored patient subgroups by the Deep Embedding with Recurrence (VaDER) model [1]. VaDER is a deep learning framework that allows for clustering of multivariate short time series with potential missingness. However, the longitudinal data in the visit-based clinical study was found to be irregular. To align disease trajectories of different patients onto a joint latent time frame, we utilized a non-linear mixed effect (NLME) model [2] to model individual disease trajectories, and shifted individual time stamps according to their random effects and fixed effects. Our hybrid model – combining NLME and VaDER approaches – achieved state-of-the-art performance. As a consequence, all the patients are grouped into two imbalanced clusters where the one with fewer people is representative of rapid disease progression (Figure 1).
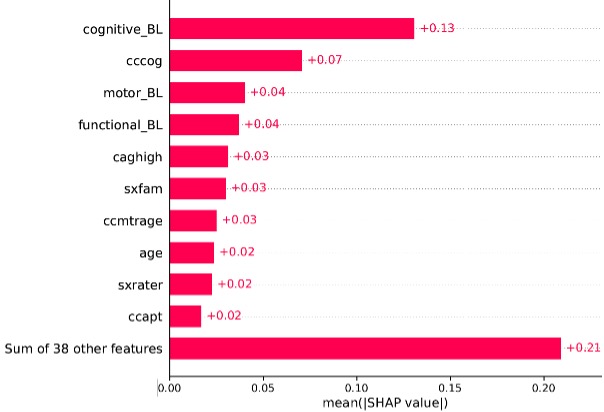
After capturing the underlying pattern of disease progression, we subsequently trained a random forest classifier that utilized cluster assignments to identify patient subgroups. The classifier facilitates the prediction of disease subtyping at an early stage and thereby indicates further disease progression. Our pipeline showed better predictive performance compared to the current approach that predicts disease onset based on only a single mutation. We did a post-hoc analysis of the feature importance for our classifier using SHAP values [3]. SHAP values explains how each feature contributes to the final prediction made by the model. The results suggest that various measures of clinical scales, age of patients’ clinical onset, as well as the length of CAG repeats in the HTT gene – Huntington’s disease is caused by its expansion – are of importance for model prediction, of which the most important one was cognitive measures (Figure 2).
In summary, our work proposed a promising data-driven pipeline that could contribute to early estimation of Huntington’s Disease progression. This pipeline is expected to expand to other neurodegenerative diseases and benefit these patients.
References:
1. de Jong, Johann, et al. "Deep learning for clustering of multivariate clinical patient trajectories with missing values." GigaScience 8.11 (2019): giz134.
2. Kühnel, Line, et al. "Simultaneous modeling of Alzheimer's disease progression via multiple cognitive scales." Statistics in Medicine 40.14 (2021): 3251-3266.
3. Lundberg, Scott M., and Su-In Lee. "A unified approach to interpreting model predictions." Advances in neural information processing systems 30 (2017).