Vinay Srinivas Bharadhwaj
Integrative analysis to identify shared mechanisms between schizophrenia and bipolar disorder and their comorbidities
PhD student Vinay Bharadhwaj talks about his recent work on identifying shared mechanisms between psychiatric disorders using knowledge-driven and gene expression data-driven analysis.
Motivation
Schizophrenia and bipolar disorder are characterized by highly similar neuropsychological signatures, implying shared neurobiological mechanisms between these two disorders. These disorders have been linked to several comorbidities, such as type 2 diabetes mellitus (T2DM) (Mukherjee et al., 1989; Argo et al., 2011). To date, an understanding of the mechanisms that mediate the link between these two disorders remains incomplete. This project aimed to gain a better understanding of the complex pathophysiology of these overlapping disorders through an integrative approach that leverages prior knowledge and experimental data.
Identification of shared mechanisms between psychiatric disorders and their comorbidities
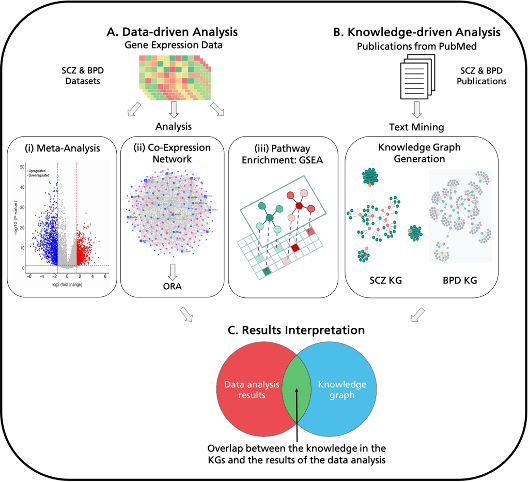
In this project, we carried out a multi-modal analysis where the gene expression data from patients with psychiatric disorders was analyzed using three different analyses. We carried out meta-analysis of the datasets using a vote-counting approach, where we identified the commonly up- and down-regulated genes across all the gene expression datasets. Next, we conducted a co-expression network analysis to identify common edges between the co-expression networks of all the other patient datasets. Finally, gene set enrichment analysis which was used to determine the dysregulated pathways using the gene expression dataset. Furthermore, schizophrenia and bipolar disorder knowledge graphs were utilized with the co-expression network analysis to identify which edges from the shared co-expression network were also available in the knowledge graph. Using this multi-modal analysis, we were able to identify several shared candidate genes and pathways between schizophrenia and bipolar disorder. Particularly, pathways related to the immune system emerged in this analysis, such as the TNF signaling pathway, IL-17 signaling pathway and NF-kappa B signaling pathway and nervous systems, such as dopaminergic synapse and GABAergic synapse. We propose that these pathways mediate the link between schizophrenia and bipolar disorder and its shared comorbidity, T2DM. Our research stands out for utilizing multiple datasets to identify mechanistic similarities between psychiatric disorders and T2DM using consensus approaches. Additionally, we incorporate prior knowledge in the form of knowledge graphs to harmonize the various approaches and obtain a more accurate assessment of shared mechanisms.
References:
- Mukherjee, S., Schnur, D. B., and Reddy, R. (1989). Family history of type 2 diabetes in schizophrenic patients. Lancet (London, England), 1(8636), 495.
- Argo, T., Carnahan, R., Barnett, M., Holman, T. L., and Perry, P. J. (2011). Diabetes prevalence estimates in schizophrenia and risk factor assessment. Annals of clinical psychiatry: official journal of the American Academy of Clinical Psychiatrists, 23(2), 117–124.