Negin Babaiha
Integrating natural language processing pipelines into causal biomedical knowledge graphs
PhD candidate Negin Babaiha talks about her work on integrating natural language processing pipelines into causal biomedical knowledge graphs.
Biomedical text mining, knowledge graphs, and their extension
One of the most difficult challenges in biomedical information extraction and literature mining is the development of effective methods and strategies for transforming scientific free text into well-structured knowledge representations such as knowledge graphs. Biomedical knowledge graphs are widely employed to support drug discovery and drug repurposing applications [1]. Inherently, knowledge evolves rapidly in many popular research domains, and therefore an efficient update procedure is in high demand for the maintenance of knowledge graphs.
The trend toward automated knowledge graph updating systems
Biomedical knowledge graphs can be built using different strategies such as from manually curated databases where the pre-existing data can be merged into a graph. Manual construction of knowledge graphs requires relevant experts to annotate and identify relevant relationships or concepts of interest and usually results in high accuracy at the expense of time. Given that manual curation is a labor-intensive task, there is an increasing trend toward semi-automatic and automatic approaches for the extension of knowledge graphs. For this reason, recent research has fast shifted to the exploration and development of text-mining tools to extract entities, scientific concepts, relationships, claims, and experimental evidence in the biomedical domain.
Introducing a system based on natural language processing (NLP) for the extension of causal biomedical knowledge graphs
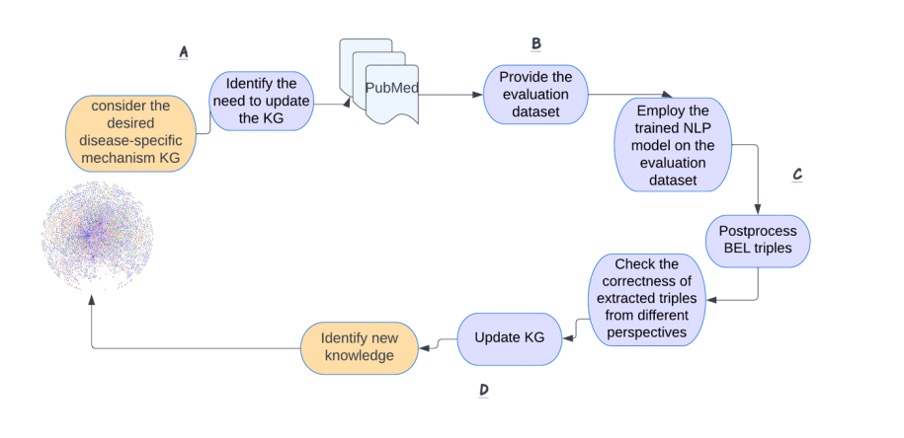
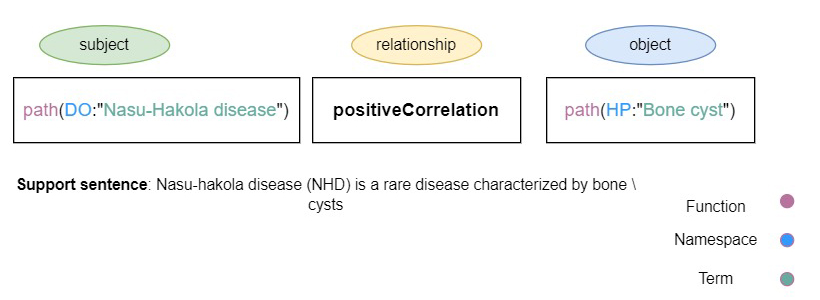
To facilitate the maintenance and updating of knowledge graphs, we aim to show the efficiency of a BERT-based NLP-based pipeline and introduce a relation extraction module. In this use case, we are studying the subgraph of Human Brain Pharmacome (HBP) that was previously curated by Lage-Rupprecht et al. [3]. This is a highly curated knowledge graph around Tau protein and its phosphorylation and has been encoded in the Biological Expression Language (BEL) [4]. BEL was developed to assemble causal relationships among biological entities with the underlying support evidence including context-specific metadata (Figure 1). Our approach is based on a pragmatic methodology to rapidly generate biomedical corpora enriched with recent publications, which are then fed into an NLP workflow for BEL triple detection. We also aim to identify previously unknown modulators of pTau posttranslational modification that are highly relevant for drug repurposing in neurodegenerative diseases. The workflow of our study for updating biomedical causal knowledge graphs is shown in Figure 2. Our strategy begins by searching PubMed for the relevant keywords. The result of this search consists of updating corpora. We then employ the NLP-based BEL triple extraction module on the updating corpora and extract relationships from it. Domain experts then analyze the extracted triples for correctness and completeness and identify the novel triples to be added to the knowledge graph. Our work is thus one of the first to explore Natural Language Processing pipelines to extend the disease-specific biomedical knowledge graph by extracting the BEL statements from biomedical publications. This work is planned to be submitted for publication soon. Stay tuned, there is more to come!
References:
1. S. Bonner et al., “Understanding the performance of knowledge graph embeddings in drug discovery,” Artif. Intell. Life Sci., vol. 2, p. 100036, Dec. 2022, doi: 10.1016/j.ailsci.2022.100036.
2. “Human Brain PHARMACOME,” Fraunhofer Institute for Algorithms and Scientific Computing SCAI. https://www.scai.fraunhofer.de/en/projects/Human-Brain-Pharmacome.html (accessed Oct. 13, 2022).
3. V. Lage-Rupprecht et al., “A hybrid approach unveils drug repurposing candidates targeting an Alzheimer pathophysiology mechanism,” Patterns, vol. 3, no. 3, Mar. 2022, doi: 10.1016/j.patter.2021.100433.
4. “PyBEL: a computational framework for Biological Expression Language | Bioinformatics | Oxford Academic.” https://academic.oup.com/bioinformatics/article/34/4/703/4557184 (accessed Oct. 04, 2022).