Astghik Sargsyan
The COVID-19 Ontology, its use case in text mining and annotation of the COVID-19 Knowledge Space
Astghik Sargsyan presents her work on the COVID-19 Ontology, describing the applications.
Motivation
Starting from late 2019, the scientific world became focused on overcoming the global COVID-19 pandemic. Even though we are involved in the research area of neurodegenerative diseases, we could not stay apart from this issue affecting everyone around the world. In order to support information organisation and harmonisation, we assembled the COVID-19 Ontology, which helps clinicians and researchers to better organize and extract information from scientific publications.
Organisation and applications of the COVID-19 Ontology
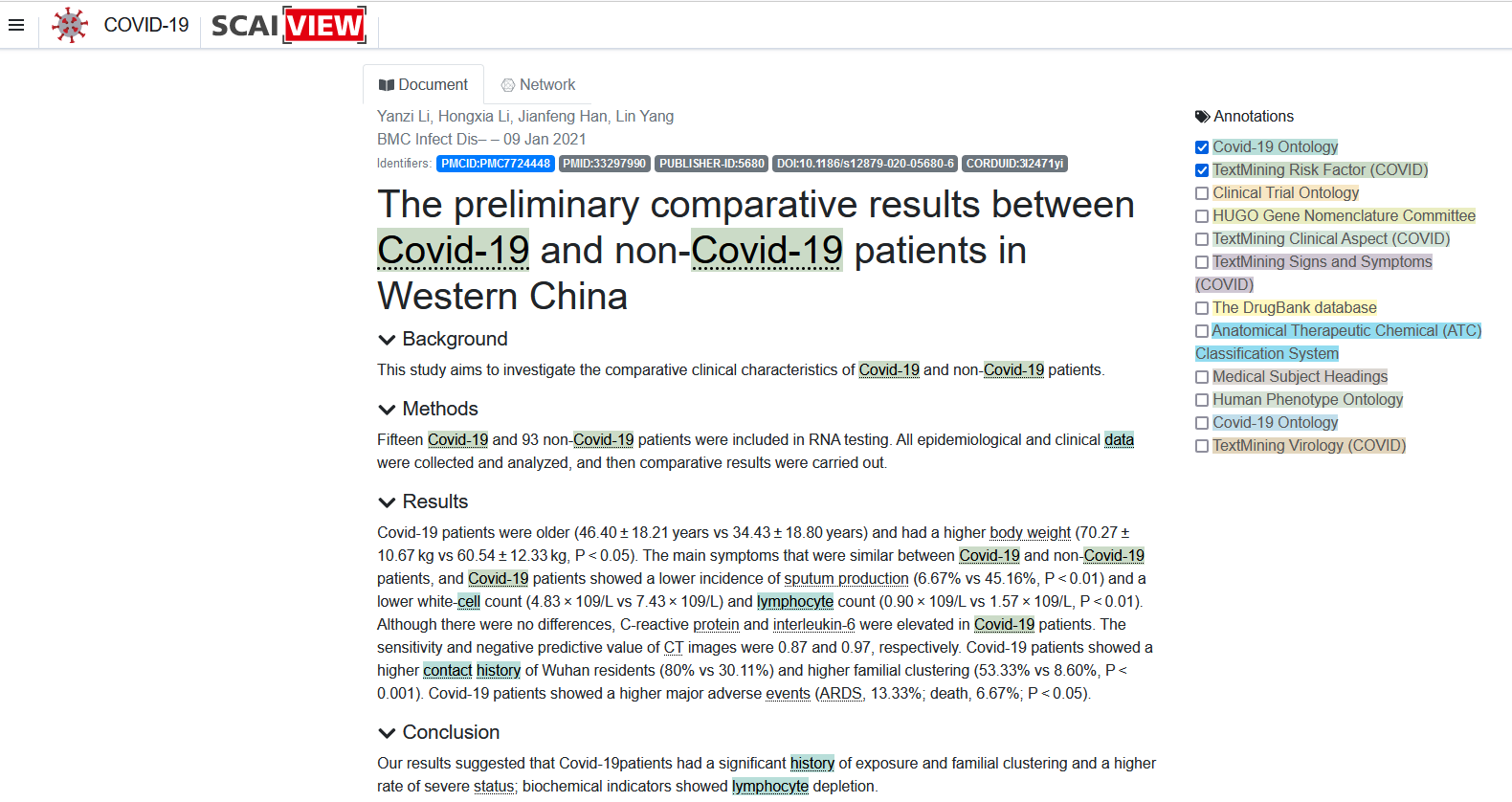
The COVID-19 Ontology comprises 2270 terms, including 2121 terms imported from existing ontologies together with 149 newly defined terms. The ontology focuses on a wide spectrum of domain-specific topics ranging from epidemiology (e.g. risk factors, transmission) via clinical aspects (e.g., signs and symptoms, diagnostics and medical intervention) and aspects of prevention and control. Other major topics include information on clinical trials, genetic and molecular processes (of both human and virus) as well as signaling pathways. In September, we published the paper on ‘The COVID-19 Ontology’ [1] and constructed it based on the Open Biological and Biomedical Ontology (OBO) principles and aligned these principles with the Basic Formal Ontology (BFO) hierarchy. The ontology itself is successfully integrated into the SCAIView engine which helps to find research- and topic- specific information.
The ontology has been successfully used for generating various research specific retrievals in SCAIView. It has also been used for various annotations of the COVID-19 Knowledge Space.
Currently the COVID-19 Ontology is being extended into a multilingual ontology, where we have translated the concepts into 14 different languages, so that the ontology could be used in various geographical regions. New concepts continue to be added to enable it for more use cases and enhanced information extraction.
-
References:
- Astghik Sargsyan, Alpha Tom Kodamullil, Shounak Baksi, Johannes Darms, Sumit Madan, Stephan Gebel, Oliver Keminer, Geena Mariya Jose, Helena Balabin, Lauren Nicole DeLong, Manfred Kohler, Marc Jacobs, Martin Hofmann-Apitius, The COVID-19 Ontology, Bioinformatics, Volume 36, Issue 24, 15 December 2020, Pages 5703–5705, https://doi.org/10.1093/bioinformatics/btaa1057
- Daniel Domingo-Fernández, Shounak Baksi, Bruce Schultz, Yojana Gadiya, Reagon Karki, Tamara Raschka, Christian Ebeling, Martin Hofmann-Apitius, Alpha Tom Kodamullil, COVID-19 Knowledge Graph: a computable, multi-modal, cause-and-effect knowledge model of COVID-19 pathophysiology, Bioinformatics, Volume 37, Issue 9, 1 May 2021, Pages 1332–1334, https://doi.org/10.1093/bioinformatics/btaa834