Avani Bhat
Development and Implementation of Auto Curation Strategies for Indication-Wide Knowledge Graphs
Avani Bhat shares a digest of her Master's thesis about a layered validation strategy to ensure that both curated and automated graphs reflect trustworthy biological knowledge.
Knowledge Graphs (KGs) have become central to biomedical research, offering structured ways to represent relationships between entities like genes, proteins, compounds, and biological processes. These graphs support critical applications in drug repurposing, disease modeling, and biomarker discovery [1].
Recently, there has been a sudden wave of large-scale, automatically generated biomedical KGs—many created using text mining or large language models (LLMs). While these models enable rapid graph construction, they also introduce risks related to data quality, hallucinations, and a lack of biological context [2]. This explosion of new KGs has made manual validation increasingly unfeasible due to the sheer volume and complexity of the data.
To address this challenge, the thesis proposes a prototype validation framework tailored to Alzheimer’s Disease KGs, helping ensure that both curated and automated graphs reflect trustworthy biological knowledge.
Methods
The validation framework was applied to two manually curated knowledge graphs – the Alzheimer’s Disease KG (12,455 triples) and the Tau KG (5,702 triples) – both developed at Fraunhofer SCAI. These were evaluated against two external KGs: KEGG Alzheimer’s KG [3] (218 triples extracted from pathway hsa05010) and Prime KG [4], a large-scale resource integrating over 4 million relationships from biomedical databases. Each graph was used selectively across the three approaches to test the framework's robustness across curated and automatically generated content.
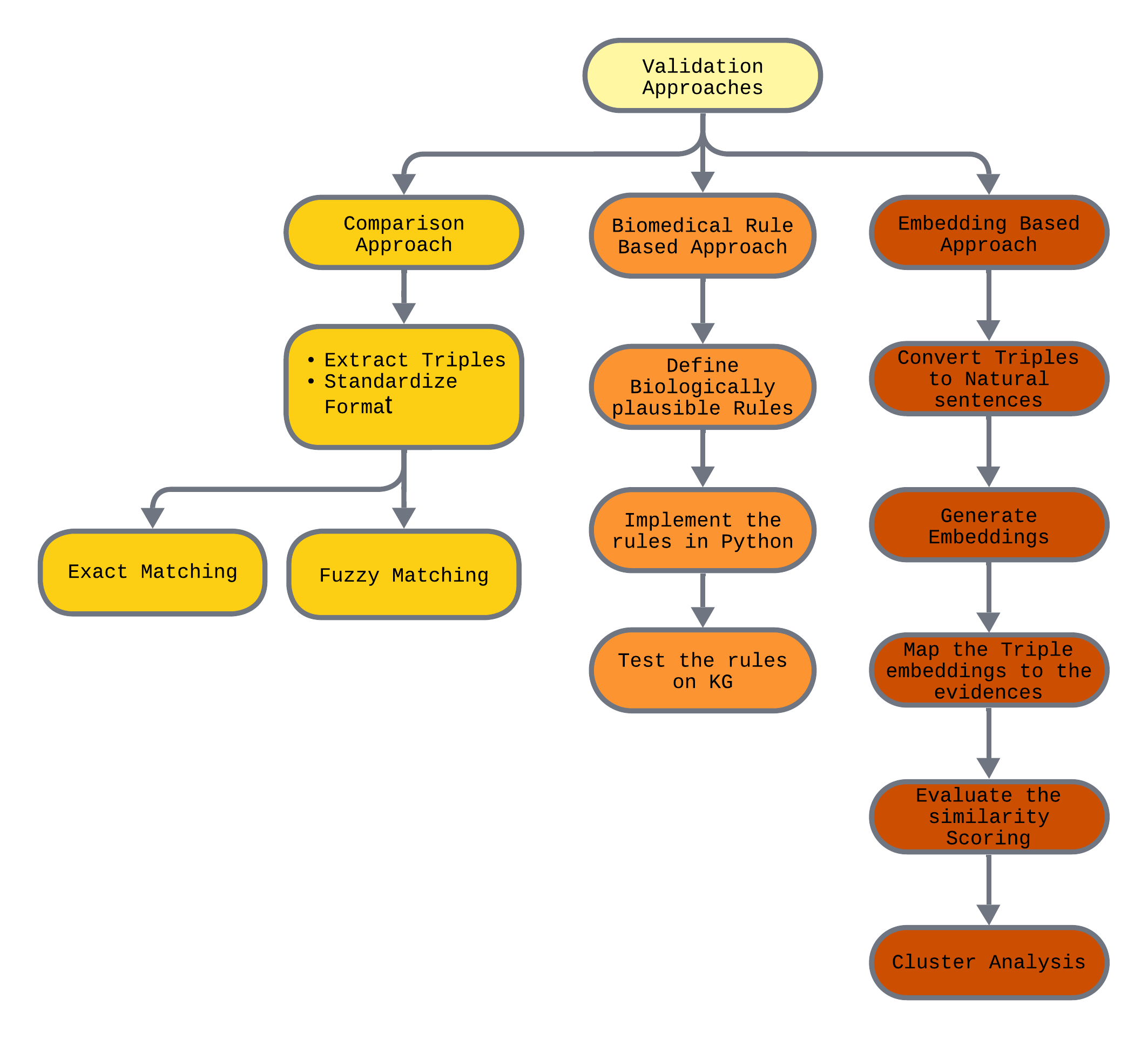
The framework (see Fig. 1) combines three distinct approaches:
1. Comparison with Curated Databases
The two manually curated Alzheimer’s Disease KGs were compared with the two trusted external sources. This comparison aimed to identify overlapping biological entities and relationships. While some similarities were found, the KGs differed in depth and detail. Instead of fuzzy matching based on entity names, triple ID-based matching using tools like eBEL could further enhance precision in such comparisons.
2. Biomedical Grammar-Based Validation
This rule-based approach involved designing grammar rules based on known biological constraints – for example, only kinases should phosphorylate proteins. These grammar rules were tested on the Tau KG and focused primarily on protein modifications (pmod), checking the plausibility of the interactions captured in the triples.
3. Embedding-Based Semantic Validation
Using OpenAI’s text-embedding-ada-002 model, triples were converted into natural language sentences and compared with their original evidence statements. This embedding-based method helped assess whether the context of the evidence aligned with the semantics of the triple, providing a scalable way to check the integrity of triple-evidence pairs.
Results and Conclusion
Each validation approach provided unique insights into the consistency and biological plausibility of the knowledge graphs:
- Comparison with External KGs (KEGG and Prime KG):
When comparing the Alzheimer’s Disease KG with KEGG, initial exact matching yielded no overlaps due to differences in predicate vocabularies. However, using fuzzy matching on just the subject-object pairs uncovered 306 shared entity pairs, suggesting that while the graphs differ structurally, they share common biological components. A similar comparison with Prime KG revealed 1,281 overlapping subject-object pairs, emphasizing the value and limitations of string-based matching techniques in identifying shared biomedical knowledge.
- Biomedical Grammar-Based Validation:
Grammar rules focused on biologically valid protein modifications (e.g., phosphorylation, acetylation, SUMOylation) were applied to the Tau KG, which includes over 1,500 modification-related triples. The validation process successfully identified ~75 plausible triples and flagged over 220 triples with inconsistencies such as non-protein subjects or incompatible modification roles, highlighting the framework's utility in filtering out biologically implausible assertions. - Embedding-Based Semantic Validation:
This approach transformed triples into natural language and compared them with evidence sentences using OpenAI embeddings. About 40% of the triples achieved high similarity scores (>0.85), showing good semantic alignment. UMAP visualizations also revealed meaningful clusters between evidence and triples, although some mismatches indicated that current LLM embeddings may fall short in handling highly technical biomedical text.
Together, these three methods offer a complementary, layered strategy for KG validation – addressing accuracy across structural, biological, and contextual dimensions. While each approach has limitations, their combination provides a promising path toward scalable and robust validation pipelines, especially as automatically generated biomedical KGs become more widespread.
Citations
[1] Zhenxing Wang and Zhongyu Wei. Pt-kgnn: A framework for pre-training biomedical knowledge graphs with graph neural networks. Computers in Biology and Medicine, 178:108768, 2024.
[2] Joan M. Boylan, Shashank Mangla, Dominic Thorn, Demian Gholipour Gha landari, Parsa Ghaffari, and Chris Hokamp. Kgvalidator: A framework for automatic validation of knowledge graph construction, arXiv:2404.15923.
[3] Kanehisa Laboratries. Kegg Alzheimer’s Knowledge Graph. https://www.kegg.jp/pathway/hsa05010, 2021.
[4] Payal Chandak, Kexin Huang, and Marinka Zitnik. Building a knowledge graph to enable precision medicine. Scientific Data, 10(1), 2022.