Lauren Nicole DeLong
Prioritization and proposition of novel COVID-19 therapies based on network representation learning
Here, Master’s student Lauren Nicole DeLong describes the work she plans to submit in which she used a network structure of protein-protein interactions combined with COVID-19 gene expression data to predict novel drug targets for COVID-19.
COVID-19: Motivating a new era of research
Following the first human infections with the Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) in late 2019 which ultimately led to the COVID-19 pandemic, many in the biomedical research community switched focus in the year 2020 to seek potential drugs, therapies, and vaccines against the virus. While multiple vaccines are now being distributed to prevent the spread of COVID-19, treatment options are still sought to attenuate symptoms and treat chronic post-COVID conditions. In addition to vaccines, the World Health Organization sees novel medications as an urgent matter to fight the ongoing COVID-19 pandemic.
Computational prediction of novel COVID-19 drug targets
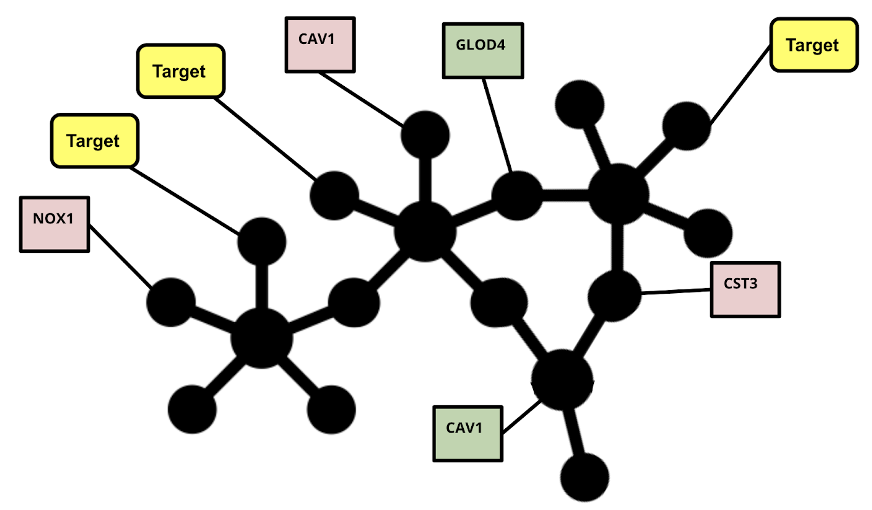
Multi-relational biomedical information about host-pathogen interactions and further disease associated genes can be modeled using graph theory by representing biological entities, like proteins, as nodes, and the relationships between them as edges. The resulting graph provides an ideal structure for SCAI’s previously published GuiltyTargets Network Representation Learning (NRL) Approach (Figure 1) [1]. Using both bulk and single-cell RNA Sequencing differential gene expression data as node features, the approach approximates similarity between nodes, or proteins, based on similar gene expression values as well as how connected they are in the graph. Then, using a simple binary classifier, the approach does its best to discriminate between the proteins in the network which are known to be potential drug targets, and those which are not based on these encoded similarity measures. Naturally, the classifier is not perfect, nor do we hope for it to be, as the information we know as humans is not necessarily ground truth. Most often, some unlabeled proteins are misclassified as potential drug targets since they are similar to the known potential drug targets, and these “false positives” serve as the predictions for novel drug targets. In addition to a binary classification, the GuiltyTargets approach ranks every protein in a lung proteome-filtered knowledge graph according to its likelihood of being a potential drug target for COVID-19 [1].
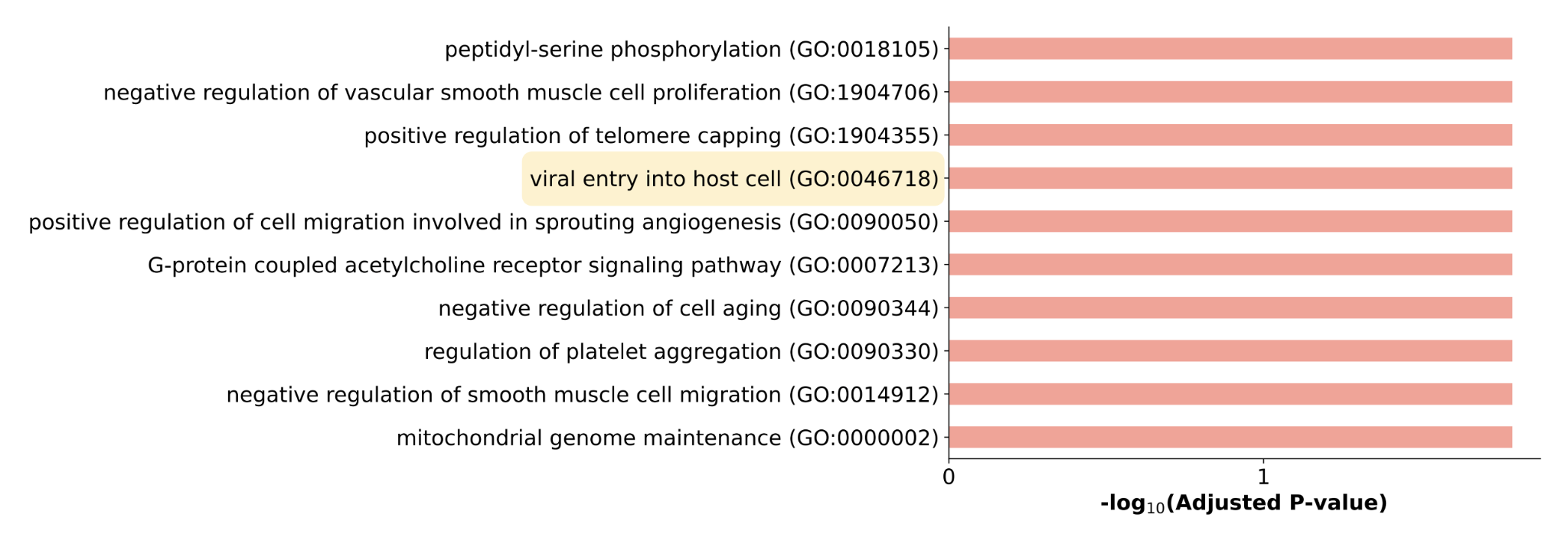
After using the method for multiple COVID-19 datasets and applying filtering steps, a list of 24 novel targets remained consistently highly ranked across multiple datasets. Of these, drugs targeting proposed targets MAP2K7, GRK2, and CBSL were experimentally validated to significantly reduce cytopathic effect of SARS-CoV-2, and drugs targeting AKT3 and PRKG1 were verified to significantly inhibit SARS-CoV-2 replication [2]. A Gene Ontology enrichment analysis revealed that this set of validated proposed targets is indeed associated with viral activity (Figure 2).
As AI-based network algorithms require sufficient data, NRL algorithms for drug target searching in the COVID-19 context are underexplored. Our work is therefore one of the first to exploit graph-based structures to uncover potential COVID-19 therapies, and we plan to submit for publication soon.
References:
- O. Muslu, C. T. Hoyt, M. P. De Lacerda,vet al. GuiltyTargets: Prioritization of Novel Therapeutic Targets with Deep Network Representation Learning. IEEE/ACM Transactions on Computational Biology and Bioinformatics, doi: 10.1109/TCBB.2020.3003830.
- Stukalov, A., Girault, V., Grass, V. et al. Multilevel proteomics reveals host perturbations by SARS-CoV-2 and SARS-CoV. Nature 594, 246–252 (2021). https://doi.org/10.1038/s41586-021-03493-4