Sepehr Golriz Khatami
Drug response simulation by calibrating patient-specific pathway signatures
In this post, Sepehr discusses his recent work on simulating drug responses in patients by calibrating their pathway activity scores using machine learning methods and a novel scoring algorithm.
Biological pathways are constructs used for organizing knowledge surrounding molecular biological processes. However, they can also be used to generate pathway activity scores for patients and subsequently train a machine learning (ML) classifier to discern between disease samples and controls. In this blog post, we present a novel method that uses such pathway activity scores to go one step beyond and employ them for drug discovery.
Our novel method operates by altering a patient’s pathway signatures so that they begin to resemble the signatures of healthy controls as a proxy for a drug’s effect. The procedure is as follows: first, we train an ML classifier to discriminate between disease samples and controls based on patient-specific pathway activity scores (Figure 1a). Next, to simulate a drug response with the help of our algorithm, we modify these scores based on pre-existing knowledge of how the drug interacts with its target(s) and the activity of the target(s) at the pathway level (Figure 1b). Finally, we pass the simulated scores to the previously trained ML model to check whether the patient sample is now predicted as a healthy control (Figure 1b). If the answer is yes for most of the patients, we consider that drug as a proxy for promising drug candidates.
To demonstrate our approach, we tested it on four different cancers with two independent drug-target datasets. Our results showed that a high proportion of the drug candidates proposed by our approach for these cancers were already FDA-approved drugs, or are currently being tested in clinical trials for the disease, beating equivalent state-of-the-art methods. If you are interested to read more about this work, check out our preprint here.
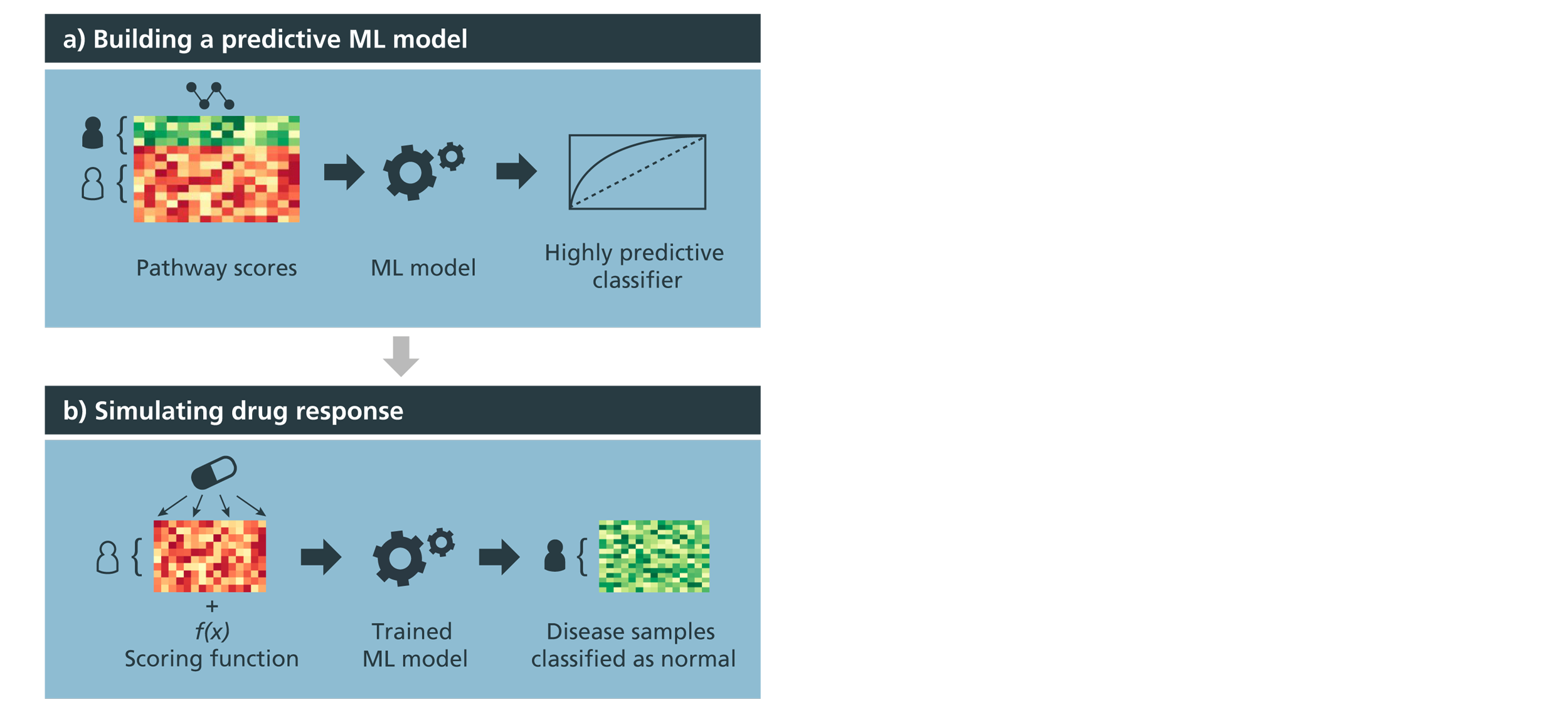
Figure 1: Conceptual overview of the drug simulation workflow and case scenario on multiple datasets.
a) Pathway activity scores are used to train a highly predictive ML model that differentiates between normal and disease samples, labelled green and red on the heatmap, respectively.
b) Next, pathway scores of disease samples are modified by using drug-target information and applying a scoring algorithm that simulates the effect of a given drug at the pathway-level. Using the modified pathway scores of disease samples, the trained ML classifier is then used to evaluate whether these modified disease samples that were previously classified as “diseased” could now be classified as “normal”.