Mission: Bringing better treatments to the right patients
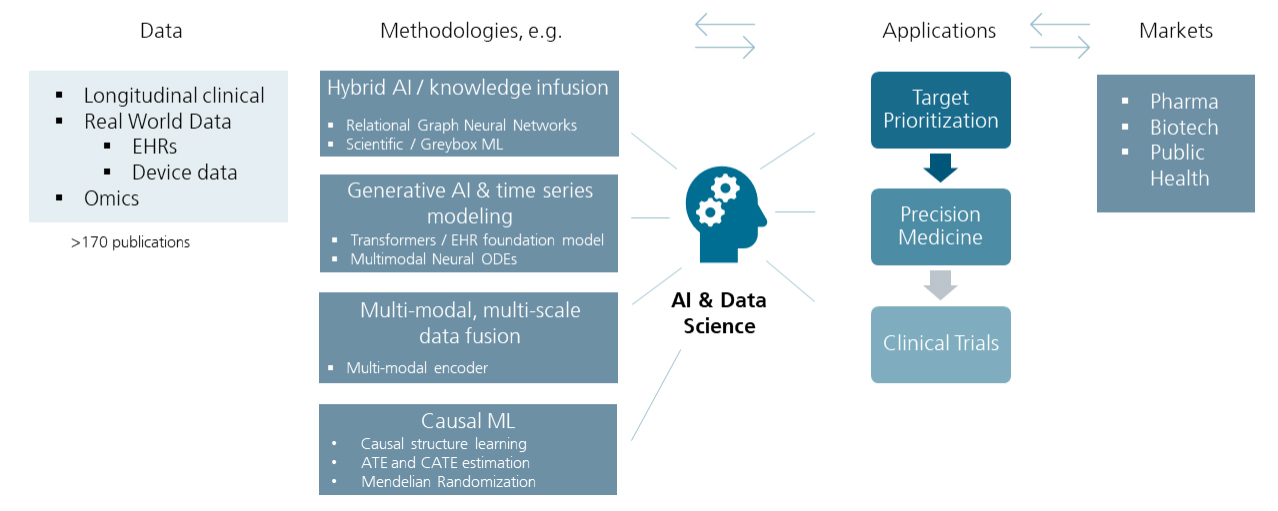
The research of the "AI & Data Science" team focuses on the development and application of AI/ML algorithms along a value chain, which largely aligns with the needs of pharma and biotech industry as well as the public health sector:
- Target prioritization (better targets):
- Rational selection of molecular target structures for future therapies
- Precision medicine (the right drug for the right patient):
- Prediction of disease risk, molecular subtype, disease progression, or treatment response at the individual patient level
- Clinical trials (better trials):
- Simulation of (counterfactual) synthetic disease trajectories
- Estimating intervention effects using real-world data
To address the highly complex issues that emerge in our applications, a broad range of AI/ML techniques is needed (covering state-of-the-art artificial neural network architectures as well as classical ML techniques). At the same time, off-the-shelf solutions often do not provide satisfactory results. Hence, a significant proportion of our work goes into the adaptation, development, and design of AI/ML approaches that are tailored to solve a particular application problem. During the last years, our work has specifically covered
- Hybrid AI/knowledge infusion
- Generative AI and time series modeling
- Multi-modal data integration
- Causal machine learning
We have a long-standing experience with a wide spectrum of relevant data types in biomedicine:
- Longitudinal clinical studies
- Real-world data:
- Electronic health records (EHRs) and claims data
- Digital markers: data derived from digital device technologies (e.g. gait sensors) and smartphone applications
- -omics